About iGMDR
iGMDR was constructed through ModelLab to collect predictive models of response for anticancer drug therapy. The models are derived from genetic analysis of cancer and its drug response experiments, and feature types of the models include common genetic variants. The models on this database not only include that have already been used in clinical practices, but also that have investigated in vivo or in vitro experiments. The construction of iGMDR should be beneficial to investigate the mechanism of action of anticancer drugs and the effectiveness of anticancer therapies, and further promote the personalized use of anticancer drugs.

iGMDR MODELS
iGMDR integrated the predictive models of anticancer drugs from various data sources such as clinical practices (e.g., FDA) and pharmacogenetic studies on cell line level (e.g., CCLE). These models were classified into 12 categories of variant types (40 subtypes), 3 types of feature levels (gene, protein, and lineage), and different grades. Each model corresponds to a Reference ID. The model-related genetic characteristics ranged from precise to specific mutation sites in a gene (e.g., SNV(ABL1 T315I)) to rough to the whole gene (e.g., MUT(ABL1)). In addition, the models we collected included not only those related to drug sensitivity and drug resistance but also those that did not respond (drug unresponsive), because we thought this would be valuable information for clinical practice or research. It is worth mentioning that the model-associated trials were for many cancer types of a certain tissue type, so the tissue type was defined and the cancer type was undefined (cancer: undefined); the model-associated trials were for all cancer types (Pan-cancer), so the tissue type was undefined (tissue: undefined). As shown in the following:
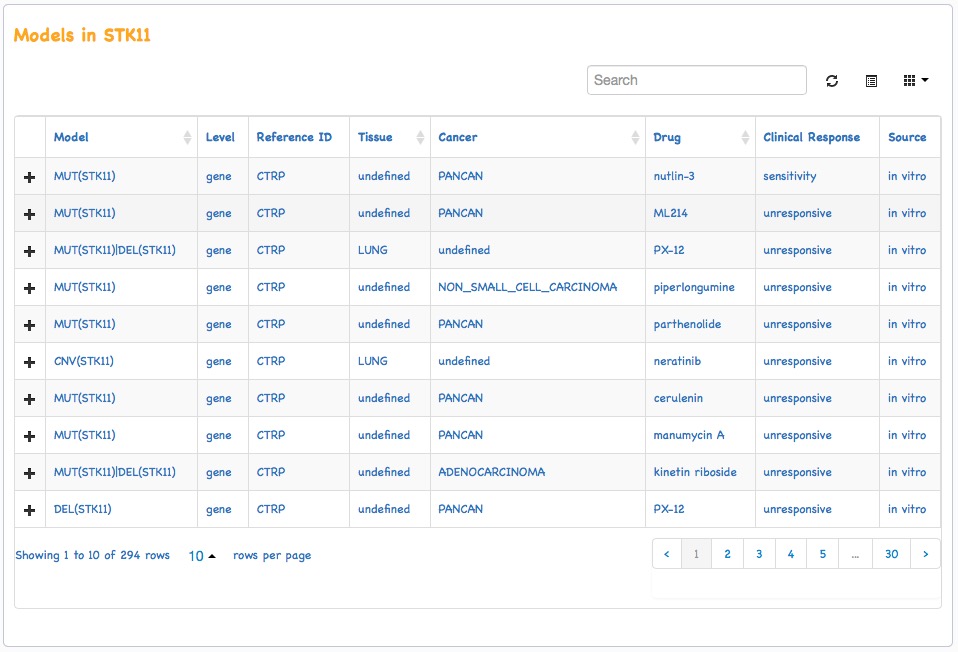
NOTE: The same event of variation, the same drug, the same tissue type, and the same cancer type, doesn't mean that the data is redundant or wrong, because different models came from different data sources and got it in different approaches. So even though there is "overlap" between the data sets of these sources, the meaning is different.
iGMDR GENES
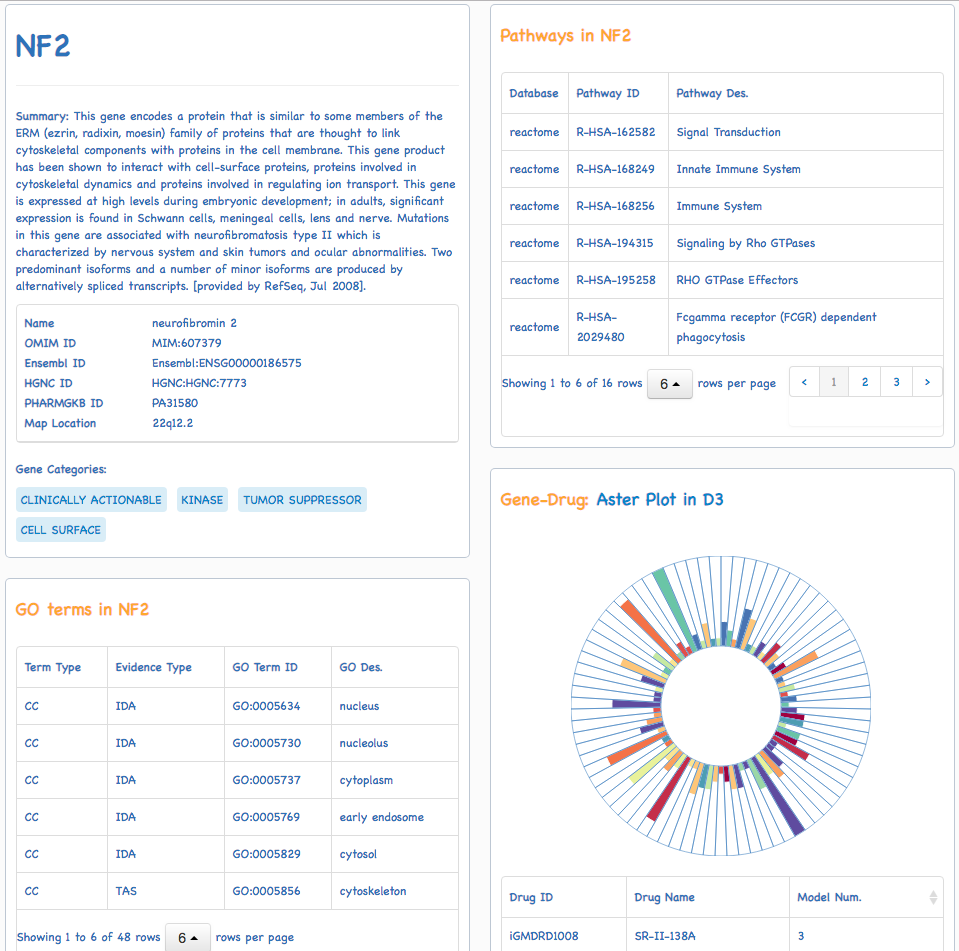
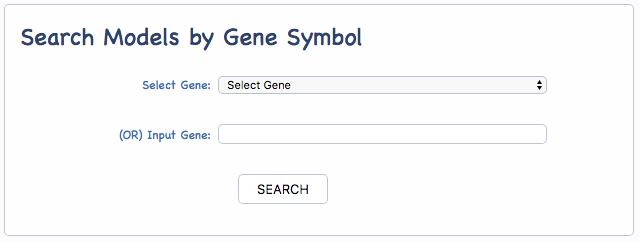
The predictive models of the anticancer drug in therapeutic response collected by iGMDR covered 4420 genes including oncogenes, tumor suppressor genes, and genes targeted by anticancer drugs in cancer-related signaling pathways. All gene names refered to HGNC (https://www.genenames.org/). In addition, gene ID (NCBI Entrez ID) was used as the index to facilitate data processing in the database table. iGMDR gene names and ids were used for model searching, and users can use the searching function in two ways: the drop-down menu of all genes or direct input a gene name or gene id. As shown in the following:
iGMDR DRUGS
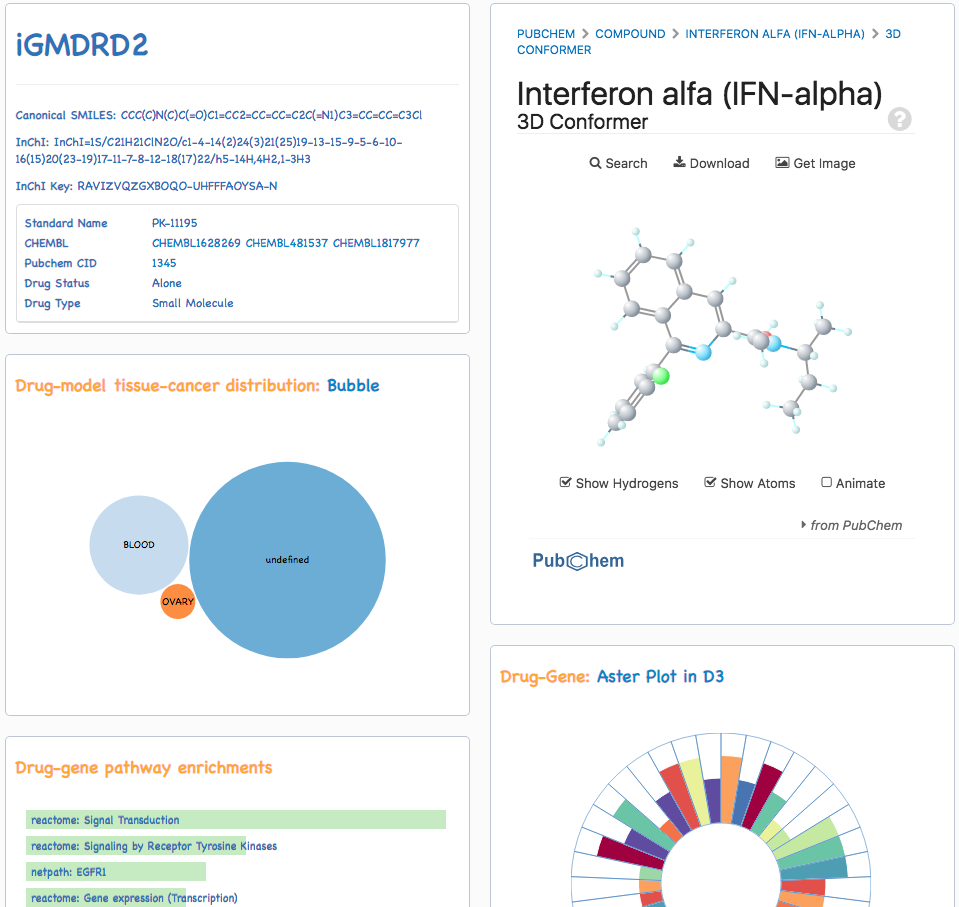
The predictive models of drug response to anti-cancer therapies collected by iGMDR involved 1,040 drugs and drug combinations. The names of drugs from different databases vary greatly. In order to manage data generally, the drug descriptions were manually standardized with the external databases. In addition, iGMDR drugs were uniformly adopted with drug id (iGMDRD) for improving data processing. iGMDR used drug names and ids as the other way to find drug-associated models. The models of drug response could not only search by the drop-down menu of all drugs but also enter a name or id of the drug directly:
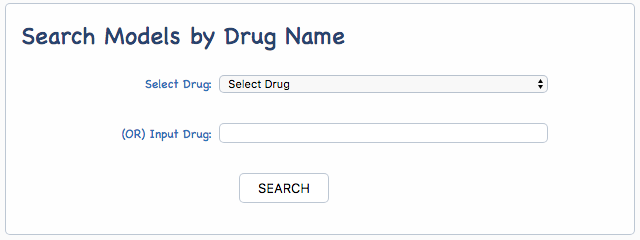
iGMDR TISSUES
As seen below, Oncotree (http://www.oncokb.org/) classified cancer types and associated them with cancer tissues. To facilitate the application of tissue information in iGMDR, Oncotree was used as a standardization of tissue type for the models. It's worth mentioning that none of the original data we collected had provided information on the tissue type. Oncotree API is as follows:
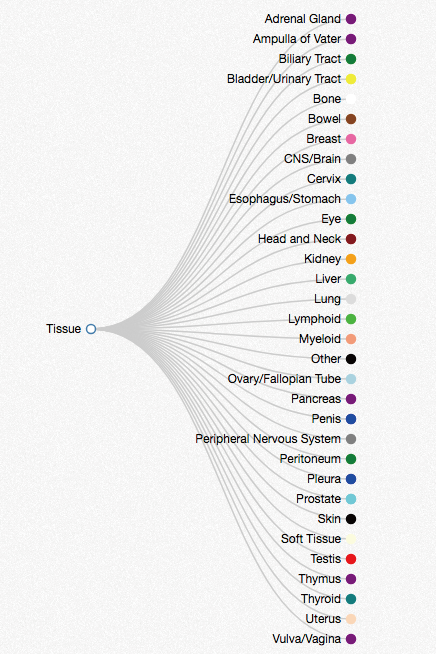
iGMDR CANCER
The names of cancer types in different resources were different. As mentioned above, the Oncotree (http://www.oncokb.org/) was introduced to manually calibrate the names of all cancer types promoting structured data storage and use.
iGMDR DATASETS
iGMDR integrated the predictive models from datasets including FDA, NCCN, CCLE, GDSC, and large-scale pharmacogenetic studies. Among them, the models from such as FDA and NCCN had been verified in clinical practises, while such as CCLE and GDSC were large-scale pharmacogenetic studies on cancer cell lines.
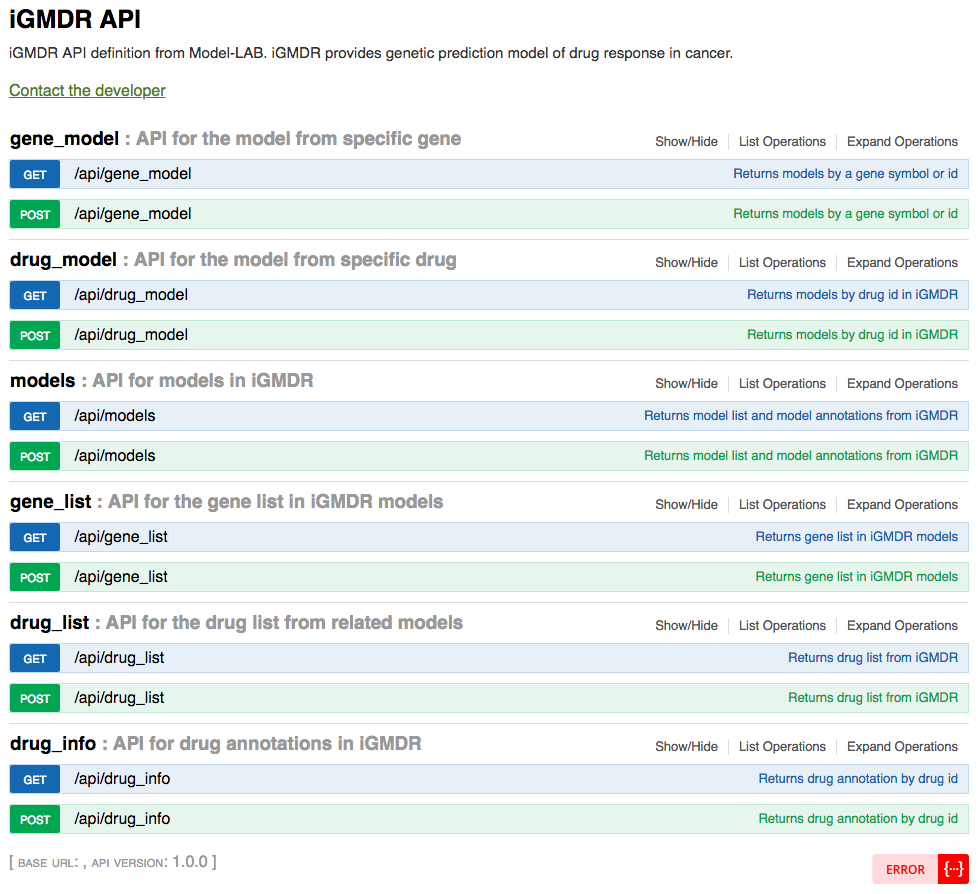
iGMDR API
iGMDR API was built on the swagger platform using PHP. iGMDR API could search model based on gene name or id and drug id, and also provided the extraction of the model list.
iGMDR CLASSIFICATION
Drug classification, target information, and signaling pathways can help users to understand the mechanism of drug action. The information of these classifications were integrated by manual collecting from the original data sources or other related databases. In addition, the external databases were correlated to improve data interaction.

